Note
Go to the end to download the full example as a Python script or as a Jupyter notebook..
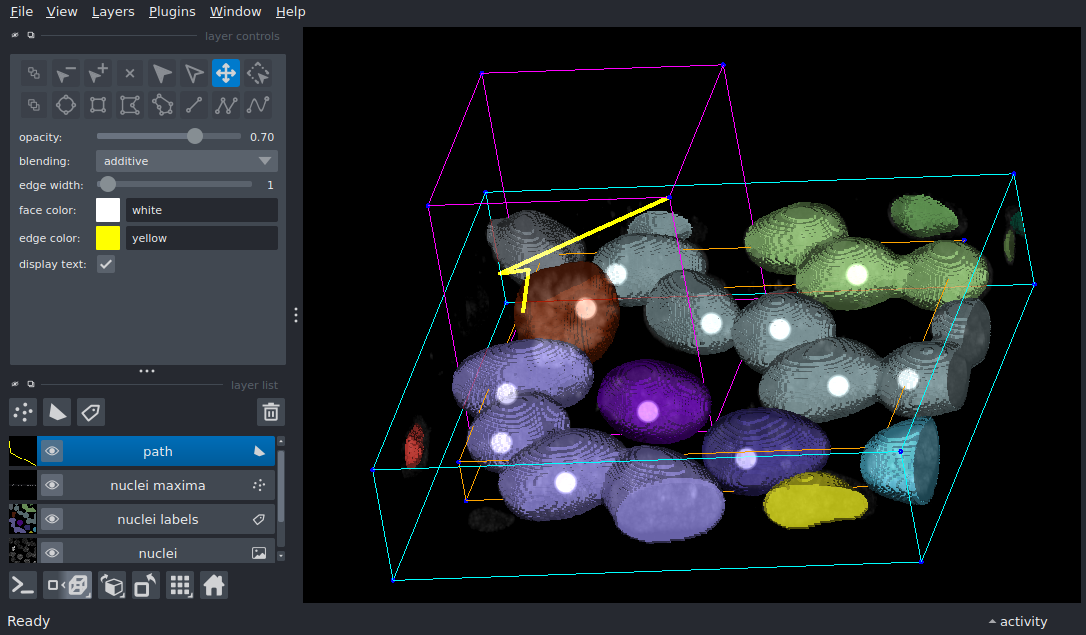
3D Layer Bounding Box Overlay#
Display multiple layer types in 3D and add a bounding box overlay to each layer.
The bounding box overlay is a visual representation of the extents of each layer. Image and label layers have data extents that are the same as the image data. Shapes and points layers have data extents that represent the convex hull of the data.
For an example showing how to modify the properties of the bounding box overlay, see Layer Bounding Box (Border) Overlay.
import numpy as np
from skimage import data, feature, filters, morphology
import napari
# get sample 3D image data, threshold, and find maxima
cells3d = data.cells3d()
nuclei = cells3d[:, 1]
nuclei_smoothed = filters.gaussian(nuclei, sigma=5)
nuclei_thresholded = nuclei_smoothed > filters.threshold_otsu(nuclei_smoothed)
nuclei_labels = morphology.label(nuclei_thresholded)
nuclei_points = feature.peak_local_max(nuclei_smoothed, min_distance=20)
# create an arbitrary path for display
path = np.array([
[0, 10, 10],
[20, 5, 15],
[56, 70, 21],
[127, 127, 127]
])
# create the viewer and display the different layer types
viewer = napari.Viewer()
viewer.add_image(nuclei, name='nuclei', contrast_limits = (10000, 65355))
viewer.add_labels(nuclei_labels, name='nuclei labels')
viewer.add_points(
nuclei_points, name='nuclei maxima', blending='additive', opacity=0.5
)
viewer.add_shapes(
path, name='path', shape_type='path', blending='additive', edge_color='yellow'
)
# add a bounding box overlay to each layer, then change the color
for layer in viewer.layers:
layer.bounding_box.visible = True
viewer.layers['nuclei labels'].bounding_box.line_color = 'cyan'
viewer.layers['nuclei maxima'].bounding_box.line_color = 'orange'
viewer.layers['path'].bounding_box.line_color = 'magenta'
# set the view to 3D and rotate camera
viewer.dims.ndisplay = 3
viewer.camera.angles = (-27, 8, -58)
if __name__ == '__main__':
napari.run()
Total running time of the script: (0 minutes 14.471 seconds)