Note
Go to the end to download the full example as a Python script or as a Jupyter notebook..
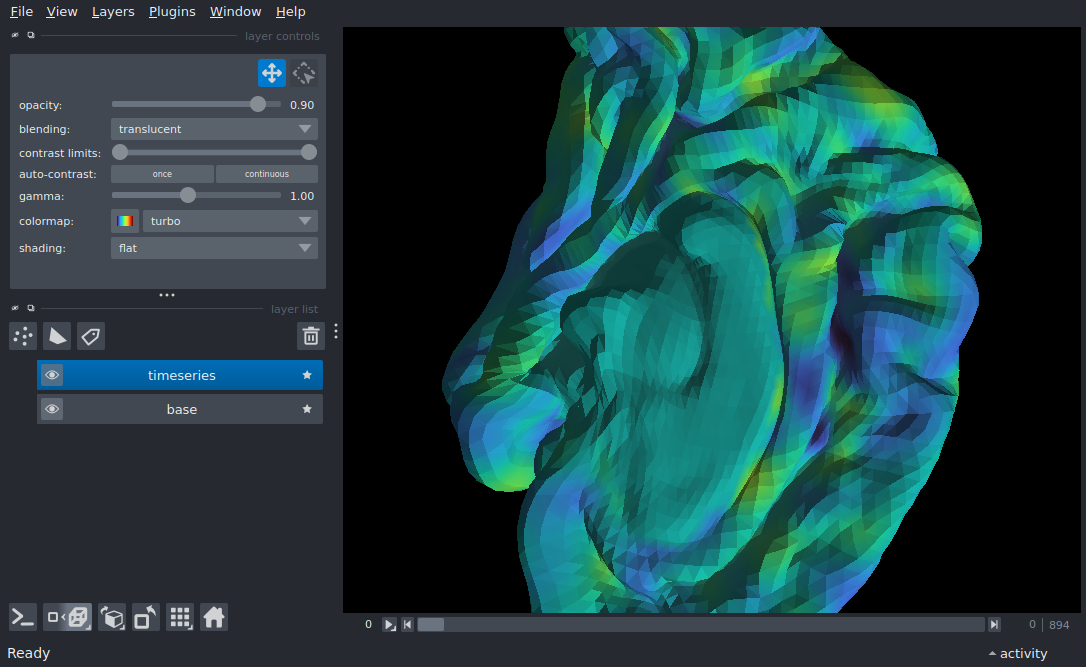
Surface timeseries#
Display a surface timeseries using data from nilearn
[fetch_surf_nki_enhanced] Added README.md to /home/runner/nilearn_data
[fetch_surf_nki_enhanced] Dataset created in
/home/runner/nilearn_data/nki_enhanced_surface
[fetch_surf_nki_enhanced] Downloading data from
https://www.nitrc.org/frs/download.php/8470/pheno_nki_nilearn.csv ...
[fetch_surf_nki_enhanced] ...done. (0 seconds, 0 min)
[fetch_surf_nki_enhanced] Downloading data from
https://www.nitrc.org/frs/download.php/8261/A00028185_rh_preprocessed_fsaverage5
_fwhm6.gii ...
[fetch_surf_nki_enhanced] ...done. (0 seconds, 0 min)
[fetch_surf_nki_enhanced] Downloading data from
https://www.nitrc.org/frs/download.php/8260/A00028185_lh_preprocessed_fsaverage5
_fwhm6.gii ...
[fetch_surf_nki_enhanced] ...done. (0 seconds, 0 min)
from importlib.metadata import version
from nilearn import datasets, surface
from packaging.version import parse
import napari
if parse(version("numpy")) >= parse('1.24') and parse(version("nilearn")) < parse('0.10.1'):
raise RuntimeError(
'Incompatible numpy version. '
'You must have numpy less than 1.24 for nilearn 0.10.1 and below to '
'work and download the example data'
)
# Fetch datasets - this will download dataset if datasets are not found
nki_dataset = datasets.fetch_surf_nki_enhanced(n_subjects=1)
fsaverage = datasets.fetch_surf_fsaverage()
# Load surface data and resting state time series from nilearn
brain_vertices, brain_faces = surface.load_surf_data(fsaverage['pial_left'])
brain_vertex_depth = surface.load_surf_data(fsaverage['sulc_left'])
timeseries = surface.load_surf_data(nki_dataset['func_left'][0])
# nilearn provides data as n_vertices x n_timepoints, but napari requires the
# vertices axis to be placed last to match NumPy broadcasting rules
timeseries = timeseries.transpose((1, 0))
# create an empty viewer
viewer = napari.Viewer(ndisplay=3)
# add the mri
viewer.add_surface((brain_vertices, brain_faces, brain_vertex_depth), name='base')
viewer.add_surface((brain_vertices, brain_faces, timeseries),
colormap='turbo', opacity=0.9,
contrast_limits=[-1.5, 3.5], name='timeseries')
if __name__ == '__main__':
napari.run()