Contributing Documentation#
This guide will teach you how to contribute to napari’s documentation.
To begin contributing, you will need:
Choose an approach to contributing:
Contributing without a local setup: This is a simpler approach that allows you to make small changes to the documentation without needing to set up a local environment. This approach is useful for quick edits or if you are not familiar with Git or the command line.
Contributing with a local setup: This is the recommended approach if you are making significant changes to the documentation, or if you want to preview your changes before submitting them. This approach requires a local setup of the napari and docs repositories.
Ask for guidance#
If you’d like to contribute a brand new document to our usage section, it’s worth opening an issue on our repository to discuss the content you’d like to see and get some early feedback from the community. The napari team can also suggest what type of document would be best suited, and whether there are already existing documents that could be expanded to include your proposed content.
Organization of the documentation#
The napari documentation is built from multiple sources that are organized into repositories:
The main napari documentation is built from sources located at the napari/docs repository on GitHub. That repository is where all the narrative documentation (e.g. tutorials, how-to guides, etc. on napari.org) pull requests should be made. This narrative napari documentation is written in MyST markdown, a version of commonmark Markdown (see a markdown cheatsheet) with some additional features.
Some of the documentation about plugins resides in the napari/npe2 repository (e.g. the contributions reference page) and should be modified there. This is documentation is written with Jinja.
The examples gallery is generated from Python source files and changes to the gallery should be made to the napari/napari repository (see also the guide on adding examples to the gallery).
Changes to how napari.org appears site-wide are made to the napari-sphinx-theme.
The API reference documentation is autogenerated from the napari source code docstrings. Docstrings are written in the reStructuredText format and any modifications to them should be submitted to the napari/napari repository.
Importantly, all of the dependencies for building the documentation are in the the
pyproject.tomlof napari/napari under the optional dependenciesdocsandgallery.
What types of documents can I contribute?#
Go to the napari/docs repo to find examples of documents you might
want to contribute. The paths are listed in parentheses below.
Explanations (in
napari/docs/guides): in depth content about napari architecture, development choices and some complex featuresTutorials (in
napari/docs/tutorials): detailed, reproducible step by step guides, usually combining multiple napari features to complete a potentially complex taskHow-tos (in
napari/docs/howtos): simple step by step guides demonstrating the use of common featuresGetting started (in
napari/docs/tutorials/fundamentals): these documents are a mix of tutorials and how-tos covering the fundamentals of installing and working with napari for beginners
The Examples gallery sources are in the main napari/napari repository
and show code examples of how to use napari.
Plugin documentation
Some of the source files for the Plugin documentation are
autogenerated from sources in the napari/npe2
repository. Any edits should be made in the napari/npe2 repo, and not to the
copy you may have in the napari/docs folder. These files’ names start with the
_npe2_ prefix and are located under the docs/plugins folder. They will be
generated when the documentation is built and the
prep_docs.py
script is run. See our
Makefile for more details.
Got materials for a workshop?
If you already have teaching materials e.g. recordings, slide decks or Jupyter notebooks hosted somewhere, you can add links to these on our napari workshops page.
Formats and templates#
Our goal is that all tutorials and how-tos are easily downloadable and executable by our users. This helps ensure that they are reproducible and makes them easier to maintain. Jupyter notebooks are a great option for our documents, because they allow you to easily combine code and well formatted text in markdown and can be executed automatically. However, their raw JSON format is not great for version control, so we use MyST Markdown documents in our repository and on napari.org.
If you are amending existing documentation, you can do so in your preferred text editor. If you wish to add a new tutorial or a how-to, we recommend you use our template. Inside the template you’ll find handy tips for taking screenshots of the viewer, hiding code cells, using style guides and what to include in the required prerequisites section.
To use the template, make a copy of docs/developers/documentation/docs_template.md
and rename it to match your content. You can edit the template directly in
Jupyter notebook, or in your preferred text editor.
Already have a notebook?
If you have an existing .ipynb Jupyter notebook that you’d like to contribute, you can convert it to MyST markdown and then edit the .md file to prepare it for contributing. For the conversion, you can install the jupytext package. Run jupytext your-notebook.ipynb --to myst to create a new MyST version of your file,
your-notebook.md. Edit this file to include the relevant sections from the docs template.
Contributing to the napari documentation without a local setup#
If you would like to see new documentation added to napari or want to see changes to existing documentation, but are not clear on these instructions or don’t have the time to write it yourself, you can open an issue.
If you are adding new documentation or modifying existing documentation and would prefer a simpler workflow than the local setup guide described below, you can use the GitHub web interface to open your pull request by either uploading file(s) from your computer, creating and editing a new file or editing an existing file on the napari/docs GitHub repository. It’s best if you first fork the napari/docs repository to your own GitHub account, create a feature branch, upload/create/edit files through the GitHub web interface, and then open a pull request from your fork back to napari-docs.
When you submit your PR, CI will kick off several jobs, including generation of a preview of
the documentation. By default, CI will use the slimfast build (“make target”), which
doesn’t build any content from outside the docs repository or run any docs notebook cells.
This is great for seeing the copy and formatting. If you want to preview other elements,
you can trigger more complete builds by commenting on the PR with:
@napari-bot make <target>
where <target> can be:
html: a full build, just like napari.org livehtml-noplot: a full build, but without the gallery examples fromnapari/naparidocs: only the content fromnapari/docs, with notebook code cells executedslimfast: the default, only the content fromnapari/docs, without code cell executionslimgallery:slimfast, but also builds the gallery examples fromnapari/napari
For more information about these targets see the “building locally” section of the documentation, including the part on specialized builds.
Once the jobs complete you will also be able to preview the documentation by
using the Check the rendered docs here! action at the bottom of your PR, which will go to a
preview website on CircleCI. Alternatively, you can download a zip file
of the build artifact and open the html files directly in your browser. For a build triggered by a comment, use
the “Triggered Docs Artifact Build” link.
If needed, a member of the maintenance team will help with updating the napari.org table of contents where necessary (by placing a reference to your new file in docs/_toc.yml) and making sure your documentation has built correctly.
Contributing to the napari documentation with a local setup#
0. Prerequisites#
Fork and clone the napari and docs repositories#
You should first fork
and then clone both the napari/napari and the napari/docs repositories to your
machine. To clone these repositories, you can follow any of the options in the GitHub guide to cloning (if you run into issues refer to the troubleshooting guide).
We recommend installing the GitHub CLI as it is easy to set up repository access permissions from the GitHub CLI and it comes with additional upside, such as the ability to checkout pull requests.
After installing the GitHub CLI you can run:
gh repo clone <your-username>/napari
gh repo clone <your-username>/docs napari-docs
Note
To reduce confusion and possible conflicts, the docs fork is being cloned into
a local repository folder named napari-docs. Alternately, you could also
rename the repository when forking napari/docs to napari-docs and then clone it via gh repo clone <your-username>/napari-docs.
Note
The napari documentation is built using make which does not work on paths which contain spaces.
It is important that you clone the napari/docs repository to a path that does not contain spaces.
For example, C:\Users\myusername\Documents\GitHub\napari-docs is a valid path, but
C:\Users\my username\Documents\GitHub\napari-docs is not.
Set up a developer installation of napari for docs building#
Because the API reference documentation (autogenerated from the napari code docstrings), the example gallery, and the documentation dependencies are sourced from the napari/napari repository, before you can build the documentation locally you will need to install from the napari/napari repository.
First, navigate to your local clone of the napari/napari repository:
cd napari
You will need:
a clean virtual environment (e.g.
conda) with Python 3.10-3.13—remember to activate it!;a from-source, editable installation of napari with the optional
docsdependencies and a Qt backend. From thenapari/naparirepository directory run, for example:python -m pip install -e ".[pyqt]" --group docs
This will use the default Qt backend. For other options, see the napari installation guide.
Note
You can combine the documentation dependencies with a development installation of napari by selecting the Qt extra and both dependency groups, e.g. installing with
.[pyqt]and adding--group dev --group docs.
Once the installation is complete, you can proceed to the directory where you cloned the napari/docs repository:
cd ../napari-docs
Here you will be able to build the documentation, allowing you to preview your document locally as it would appear on napari.org.
1. Write your documentation#
Depending on the type of contribution you are making, you may be able to skip some steps:
If you are amending an existing document you can skip straight to 3. Preview your document
For all other documentation changes, follow the steps below.
How to check for broken links
If you have modified lots of document links, you can check that they all work by running make linkcheck-files in the napari/docs folder. However, this can take a long time to run, so if you have only modified links in a single document, you can run:
make linkcheck-files FILES=path/to/your/document.md
2. Update the table of contents (TOC)#
If you are adding a new documentation file, you will need to add your document
to the correct folder based on its content (see the list above
for common locations), and update docs/_toc.yml.
If you’re adding a document
to an existing group, simply add a new - file: entry in the appropriate spot. For example, if I wanted to add
a progress_bars.md how to guide, I would place it in docs/howtos and update _toc.yml as below:
- file: howtos/index
subtrees:
- titlesonly: True
entries:
- file: howtos/layers/index
subtrees:
- titlesonly: True
entries:
- file: howtos/layers/image
- file: howtos/layers/labels
- file: howtos/layers/points
- file: howtos/layers/shapes
- file: howtos/layers/surface
- file: howtos/layers/tracks
- file: howtos/layers/vectors
- file: howtos/connecting_events
- file: howtos/napari_imageJ
- file: howtos/docker
- file: howtos/perfmon
- file: howtos/progress_bars # added
To create a new subheading, you need a subtrees entry. For example, if I wanted to add geo_tutorial1.md and geo_tutorial2.md
to a new geosciences subheading in tutorials, I would place my documents in a new folder docs/tutorials/geosciences,
together with an index.md that describes what these tutorials would be about, and then update _toc.yml as below:
- file: tutorials/index
subtrees:
- entries:
- file: tutorials/annotation/index
subtrees:
- entries:
- file: tutorials/annotation/annotate_points
- file: tutorials/processing/index
subtrees:
- entries:
- file: tutorials/processing/dask
- file: tutorials/segmentation/index
subtrees:
- entries:
- file: tutorials/segmentation/annotate_segmentation
- file: tutorials/tracking/index
subtrees:
- entries:
- file: tutorials/tracking/cell_tracking
- file: tutorials/geosciences/index # added
subtrees: # added
- entries: # added
- file: tutorials/geosciences/geo_tutorial1 # added
- file: tutorials/geosciences/geo_tutorial2 # added
3. Preview your document#
If your documentation change includes code, it is important that you ensure the code is working and executable. This is why you will need to have a development installation of napari installed. Examples are automatically executed when the documentation is built and code problems can also be caught when previewing the built documentation.
There are two ways you can build and preview the documentation website as it would appear on napari.org:
building locally - this requires more setup but will allow you to more quickly check if your changes render correctly.
view documentation in a GitHub pull request - this requires no setup but you will have to push each change as a commit to your pull request and wait for the continuous integration workflow to finish building the documentation.
Tip
To see the markdown document structure and content change in real-time without building, you can use a MyST markdown preview tool like VScode with the MyST extension or MyST live preview. This can also help you to spot any markdown formatting errors that may have occurred. However, this MyST markdown preview will have some differences to the final built html documentation due to autogeneration, so it is still important to build and preview the documentation before submitting your pull request.
3.1. Building locally#
To build the documentation locally we use the
make tool to execute
sphinx builds, as defined in the
Makefile at the root of the docs repository.
Once the build is completed rendered HTML will be placed in docs/_build/html.
Find index.html in this folder and drag it into a browser to preview the
website with your new document.
You can also run this Python one-liner to deploy a quick local server on http://localhost:8000:
$ python3 -m http.server --directory docs/_build/html
Note
The entire build process pulls together files from multiple sources and can be
time consuming, with a full build taking upwards of 20 minutes. Additionally,
building the examples gallery, as well as executing notebook cells will
repeatedly launch napari, resulting in flashing windows.
As a result, there are several partial-build options available in addition to a full build that only build certain parts of the full documentation build. Depending on what you want to contribute, you may never need to run the full build locally. For example, maybe you don’t want to build the examples gallery or you only want to edit copy and not run the notebook cells or only want to edit a single napari example. See Building what you need for details.
Running a full build#
To run a full documentation build from scratch, matching what is deployed at napari.org, run:
make html
from the root of your local clone of the napari/docs repository (assuming you’ve installed
napari with the docs prerequisites). Note this can take upwards of 20 minutes
and will repeatedly pop up napari viewers as examples and notebook cells are executed.
If the changes you have made to documentation don’t involve the gallery of napari examples,
which are in the examples directory in the napari repository, you can speed up this
build by running:
make html-noplot
This will skip the gallery build, which involves launching up napari and rendering
all the examples, but it will still build all of the other content, including the
UI architecture diagrams, events reference, and
preferences, which are generated from sources in the napari
repository. This will also run all notebook cells.
Note
The make html command above assumes you have a local clone of the
napari/napari repo at the same level as
the napari/docs clone. If that’s not the case, you can specify the location of
the examples gallery folder by executing
make html GALLERY_PATH=<path-to-examples-folder>
The GALLERY_PATH option must be given relative to the docs folder. If your
folder structure is
├── napari-docs
│ └── docs
├── napari
│ ├── binder
│ ├── examples
│ ├── napari
│ ├── napari_builtins
│ ├── resources
│ └── tools
Then the command would be
make html GALLERY_PATH=../../napari/examples
Update documentation on file change
We’ve provided several build variants with -live that will use
sphinx-autobuild.
When using these make variants, when you save a file, re-builds
of the changed file will be triggered automatically and will be faster,
because not everything will be built from scratch. Further, a browser preview
will open up automatically at http://127.0.0.1, no need for further action!
Edit the documents at will, and the browser will auto-reload.
Once you are done with the live previews, you can exit via Ctrl+C
on your terminal.
For example, if you are not editing the gallery examples in the napari repository, but otherwise want a full build, then you can use:
make html-noplot-live
The first run will be a full build (without the gallery) so a number of napari instances will pop up, but then when re-building on save only edited files will be rebuilt.
For faster reloads, you can try:
make html-live SPHINXOPTS="-j4"
Note: using -j4 will parallelize the build over 4 cores and can result in crashes.
Headless GUI builds
If you are running a full build, you can run it in a “headless GUI” mode, which will prevent napari windows from popping up during the build.
If you are using X11 as your display server and you have xvfb installed on your system, you can use the
docs-xvfbcommand:make docs-xvfbThis will prevent all but the first napari window from being shown during the docs build.
If you are using Wayland as your display server, and you have
xwfb-runinstalled on your system (part of the xwayland-run utilities, you can use thedocs-xwfbcommand:make docs-xwfbNote that you may have to install the
xwayland-runpackage manually so that it uses the correct Python environment you are using to build the docs. You can do that by cloning the sources from the xwayland-run GitLab repository and running:meson setup -Dcompositor=weston --prefix=/path/to/env/ -Dpython.install_env=prefix . build meson install -C build
Building what you need#
napari/docs and notebooks
If you only want to edit materials in the docs repository, including
notebook code cell outputs (e.g. any of the tutorials), but you don’t need any
of the sources in napari/napari built, then you can use the docs build variant. Run:
make docs
or
make docs-live
Note that this will still execute the docs repository notebook code cells,
resulting in napari windows popping up repeatedly. The -live
variant will open a browser preview and auto-rebuild any pages you edit.
napari/docs only
If you you only want to edit copy in the docs repository and don’t need any
of the sources in napari/napari built nor the notebook cell outputs, then you can use one
of the slim builds. Run:
make slim
or
make slimfast
or
make slimfast-live
These will not build any of the external content (such as the gallery) and will
not execute code cells. slimfast will run the build in parallel, which can be
significantly faster on multi-core machines (under a minute), while slimfast-live
will open a browser preview and auto-rebuild any pages you edit.
See the -live builds note.
napari/docs and napari gallery of examples
If you are working on the napari examples and want to build the whole examples
gallery, but not other external content nor the docs notebook cell outputs,
then you can use:
make slimgallery
or
make slimgallery-live
These builds will build the documentation with the entire gallery. The -live
variant will will open a browser preview and auto-rebuild any pages you edit.
napari/docs and a single example in the gallery
If you want to work on a single example Python script in the napari repository
examples directory, you can build the documentation with just a chosen example by
specifying it by name. For example, to build the vortex.py example, run:
make slimgallery-vortex
or
make slimgallery-live-vortex
This will only execute and build the single chosen example. The -live
variant will open a browser preview and auto-rebuild the single example or
any other docs pages on edit, but it will not run any other code cells.
Additional utilities in the Makefile#
To clean up (delete) generated content, including auto-generated .rst and .md files,
as well as the html files, you can use:
make clean
Running this is a good idea if you have not built the documentation in a long
time and there may have been significant changes to the docs repository, e.g.
changes to the table of contents or layout.
To clean up the files generated for the examples gallery, you can use:
make clean-gallery
Running this is a good idea if you have not built the documentation in a long
time and there may have been significant changes to the napari examples.
To generate the source files from the napari repository, including the UI architecture diagrams, events reference, and preferences, you can run:
make prep-docs
Note that this is can take upwards of 10 minutes! Run this if you are interested
in building the external resources or editing the scripts that generate them in
docs/docs/_scripts. In most cases make prep-stubs will suffice, which will
generate place-holder stub files for the external content.
3.2. View in GitHub Pull Request#
Alternatively, when you submit your pull request, the napari/docs repository continuous integration includes a GitHub action that builds the documentation and saves the artifact for you to preview or download. Note you will need to 4. Submit your pull request first. You can then view it in one of two ways:
preview on your browser via CircleCI in just one click - this is the easiest method but in rare cases it may not match the documentation that is actually deployed to napari.org.
download the built documentation artifact and view it locally - this is more complicated, but the built docs will always match what is deployed to napari.org.
When you submit a pull request to the napari/napari repository, its continuous integration will only build the docs in CircleCI. Thus you will only be able to preview the documentation on CircleCI.
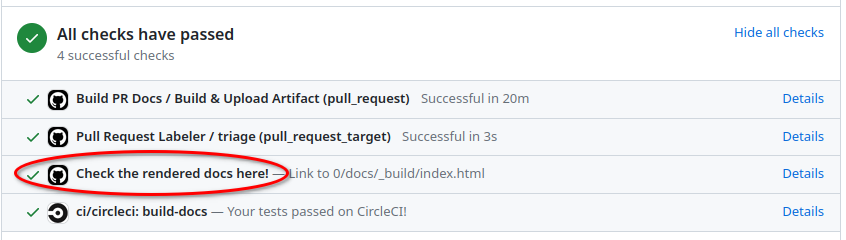
Preview on CircleCI#
Simply click on Details next to the Check the rendered docs here! at the bottom
of your pull request:
Download documentation artifact#
Click on Details next to
Build & Deploy PR Docs / Build & Upload Artifact (pull_request):
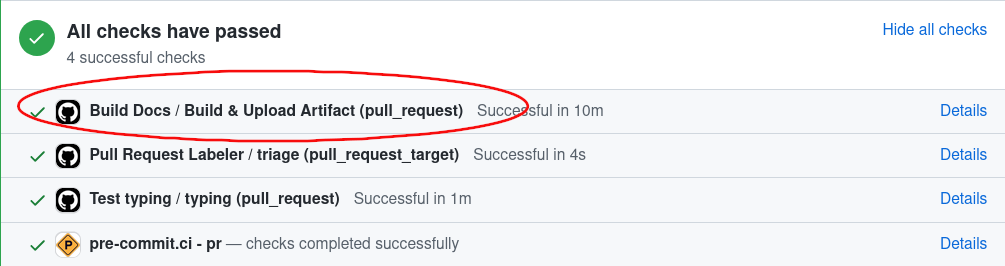
Click on Summary on the top left corner:
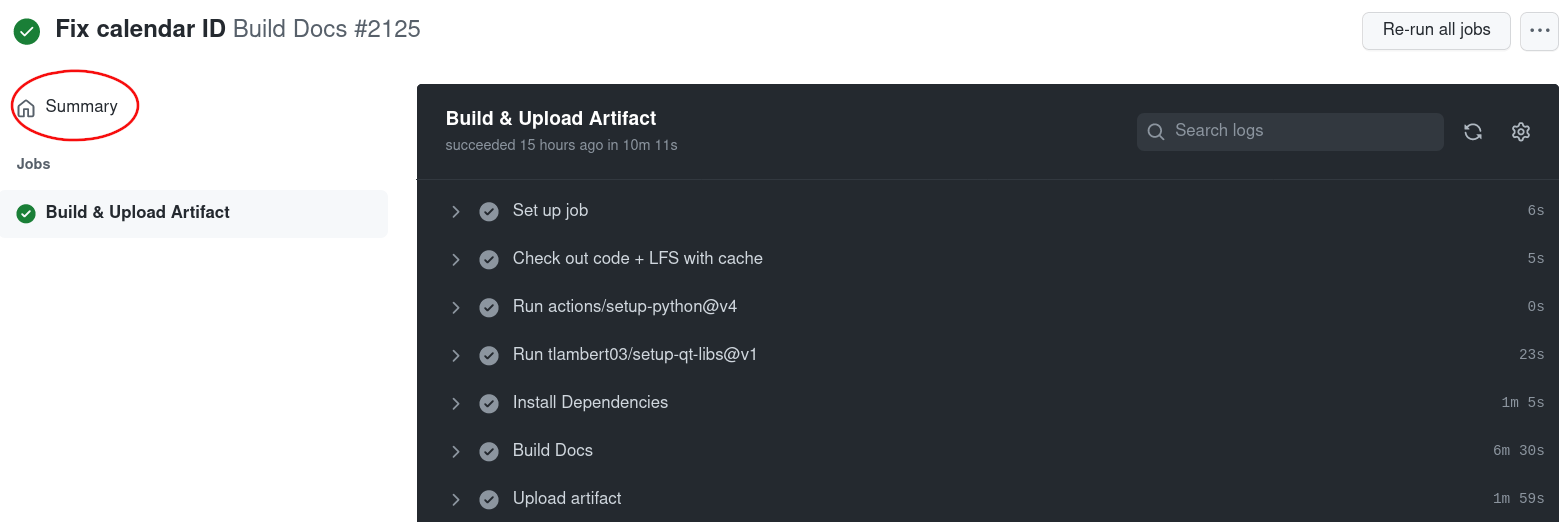
Scroll down to Artifacts and click on html to download the built documentation:
Extract the compressed archive and open the
html/index.htmlfile on your preferred browser. You can also use Python’shttp.servermodule to open a local server on http://localhost:8000:
$ cd ~/Downloads/html # `cd` to the path where you extracted the 'html' artifact
$ python3 -m http.server
4. Submit your pull request#
Once you have written and previewed your document, it’s time to open a pull request to napari’s docs repository and contribute it to our codebase.
If you are simply contributing one file (e.g., a tutorial or how-to page) you can use the GitHub web interface to open your pull request. Ensure your document is added to the correct folder based on its content (see the list above for common locations).
To open a pull request via git and the command line, follow this guide. You can also reach out to us on zulip for assistance!
Not sure where to place your document or update _toc.yml? Make a best guess and open the pull request - the napari team will
help you edit your document and find the right spot!
Building the documentation on Windows#
Note
It is very important that you clone the napari/docs repository to a path that does not contain spaces.
For example, C:\Users\myusername\Documents\GitHub\napari-docs is a valid path, but
C:\Users\my username\Documents\GitHub\napari-docs is not.
If you clone the napari-docs repository to a directory following the default Windows path naming convention, e.g.
C:\Users\my username\Documents\GitHub\napari-docs (note the space), and run the make commands to build the napari docs, it may remove unintended files from your computer as it will essentially run the command rm -rf C:\Users.
This is because the napari documentation is built using make which does not work on paths which contain spaces.
The documentation build requires some Linux specific commands, so some extra steps are required to build the documentation on Windows. There are multiple tools for this, but Git Bash or Windows Subsystem for Linux (WSL) are recommended.
Git Bash#
First, you will need to install make on Windows:
Install Chocolatey (a Windows package manager) by following the instructions here.
Install
makewithchoco install make.
Alternatively, you can download the latest make binary without guile from ezwinports and add it to your PATH.
Then install Git Bash and build the documentation:
Install Git Bash (you should already have this if you use
giton Windows).Activate your virtual environment in Git Bash.
Conda environment: To have your conda environment available in Git Bash, launch Git Bash, then run
conda init bashfrom anaconda prompt and restart Git Bash. The conda environment can then be activated from Git Bash withconda activate <env_name>.Virtualenv: To have your virtualenv available in Git Bash, launch Git Bash, then run
source <path_to_virtualenv>/Scripts/activate.
From Git Bash,
cdto the napari docs repository and runmake htmlor othermakecommands to build the documentation.
Tip
If you use Git Bash a lot, you may want to set conda to not initialize on bash by default to speed up the launch process. This can be done with conda config --set auto_activate_base false. You can then activate conda in Git Bash with conda activate base.
Note
If you are using an IDE, it is likely that it will not use Git Bash by default. You may need to configure your IDE to use Git Bash as the default terminal for the napari docs. For example, in VS Code, you can set the default terminal to Git Bash for the napari docs repository by adding the following to your workspace settings:
"terminal.integrated.defaultProfile.windows": "Git Bash"
Windows Subsystem for Linux (WSL)#
Alternatively, you can install WSL, which will allow you to run a Linux environment directly on Windows (without any virtual machines, etc.). You need to have Windows 10 version 2004 and higher or Windows 11. Then you can run scripts and command line utilities, as well as python and napari from for example Ubuntu on your Windows machine.
Install the Windows Subsystem for Linux and choose a linux distribution. We will use Ubuntu for this guide since it is the default WSL distribution, easy to install, and works well with WSLg. The default method to perform this installation is to run
wsl --install -d Ubuntufrom command prompt as an administrator but you can refer to the guide for other installation methods.Restart your computer. On restart, you will be prompted to create a user account for WSL. This account is separate from your Windows account, but you can use the same username and password if you wish.
Open up the Ubuntu distribution via the
Ubuntucommand and runsudo apt update && sudo apt upgradeto update the distribution.Install a napari development environment in Ubuntu following the contributor guide and activate the virtual environment that napari was installed into.
Install some common QT packages and OpenGL
sudo apt install -y libdbus-1-3 libxkbcommon-x11-0 libxcb-icccm4 libxcb-image0 libxcb-keysyms1 libxcb-randr0 libxcb-render-util0 libxcb-xinerama0 libxcb-xinput0 libxcb-xfixes0 mesa-utils libglu1-mesa-dev freeglut3-dev mesa-common-dev '^libxcb.*-dev' libx11-xcb-dev libxrender-dev libxi-dev libxkbcommon-dev libxkbcommon-x11-dev.You can test that all of this OpenGL setup is working by running
glxgearsfrom the Ubuntu terminal. You should see a window with some gears spinning.sudo apt install fontconfig.pip install pyqt5-tools.Fork the napari docs repository and clone it to the same parent folder as the napari repository (see 0. Prerequisites). Then navigate to the napari docs folder via
cd napari-docs.Install
makewithsudo apt install make.Run
make htmlor othermakecommands to build the documentation.
Route graphical output to Windows
By default, the graphical interface to glxgears or napari from WSL should be visible on Windows via WSLg without any configuration.
However, if you are getting errors running glxgears or can’t see the interface to graphical applications, then you may need to route the graphical output to Windows. To do this:
Install an Xserver for Windows, Vcxsrv. When launching it, choose the options as default, except tick “disable access control”.
Export environment variables (you will need to do this for every new shell you open, unless you add them to your
.bashrc):mkdir ~/temp export DISPLAY=$(awk '/nameserver / {print $2; exit}' /etc/resolv.conf 2>/dev/null):0 export LIBGL_ALWAYS_INDIRECT=0 export XDG_RUNTIME_DIR=~/temp export RUNLEVEL=3
Run
glxgearsfrom the Ubuntu terminal. You should see a window with some gears spinning.
Adding examples to the Gallery#
All of the examples in the examples Gallery are Python scripts that
live under the examples folder of the napari repository.
Because of how this gallery is built, every such Python script needs a docstring
containing at least a title (following the .rst format of docstrings in
Python) and one or more tags. This ensures the example will be shown in the
correct place in the table of contents for the gallery (see, for example, the
Tags page.)
Here’s an example of what that means. The example file add_image.py contains the following:
"""
Add image
=========
Display one image using the :func:`view_image` API.
.. tags:: visualization-basic
"""
from skimage import data
import napari
# create the viewer with an image
viewer, layer = napari.imshow(data.astronaut(), rgb=True)
if __name__ == '__main__':
napari.run()
The equals signs under “Add image” mark this as the example title. The text
below is a short description of the example, and the tags are assigned by the
use of the .. tags:: directive. In this case, the only tag associated with
this example is visualization-basic. Multiple tags can be separated with a
comma.
Note that the examples are .py source files, and any outputs and images will
be autogenerated when the documentation site is built.
Note
If you are running any of these examples in an interactive environment (e.g. a Jupyter notebook or IPython console), the block
if __name__ == '__main__':
napari.run()
is not necessary—you can copy-paste the code above it. However, it is required when running examples as scripts, using for example python add_image.py.
Because our example gallery is built from Python scripts, you need to ensure this
block is present in all contributed examples.
Finally, you can find the current example dependencies in the pyproject.toml of napari/napari under the optional dependency gallery. If you add an example with a new dependency, be sure to include it here such that your example can be properly tested and built into the documentation.
Cross-referencing Gallery examples#
If you want to generate links to Gallery examples from anywhere in the docs, then the cross-referencing format you use will depend on the format of the doc you are writing. Note that the gallery examples live in /gallery despite being in napari/examples because docs/docs/conf.py specifies that examples are built into the gallery directory. The Sphinx cross-reference namespace is generated with the sphx_glr prefix, then path separators are converted to underscores, for example /gallery/add_image.py becomes _gallery_add_image.py to get the end result sphx_glr_gallery_add_image.py.
For
.mdfiles (myst, used in the majority of docs), use the{ref}directive.{ref}`sphx_glr_gallery_add_image.py`displays as: Add imageFor
.pyfiles (rst, used in the example gallery), cross-reference with the:ref:directive::ref:`sphx_glr_gallery_example.py`.