Note
Go to the end to download the full example code
nD vectors¶
Display two vectors layers ontop of a 4-D image layer. One of the vectors layers is 3D and “sliced” with a different set of vectors appearing on different 3D slices. Another is 2D and “broadcast” with the same vectors apprearing on each slice.
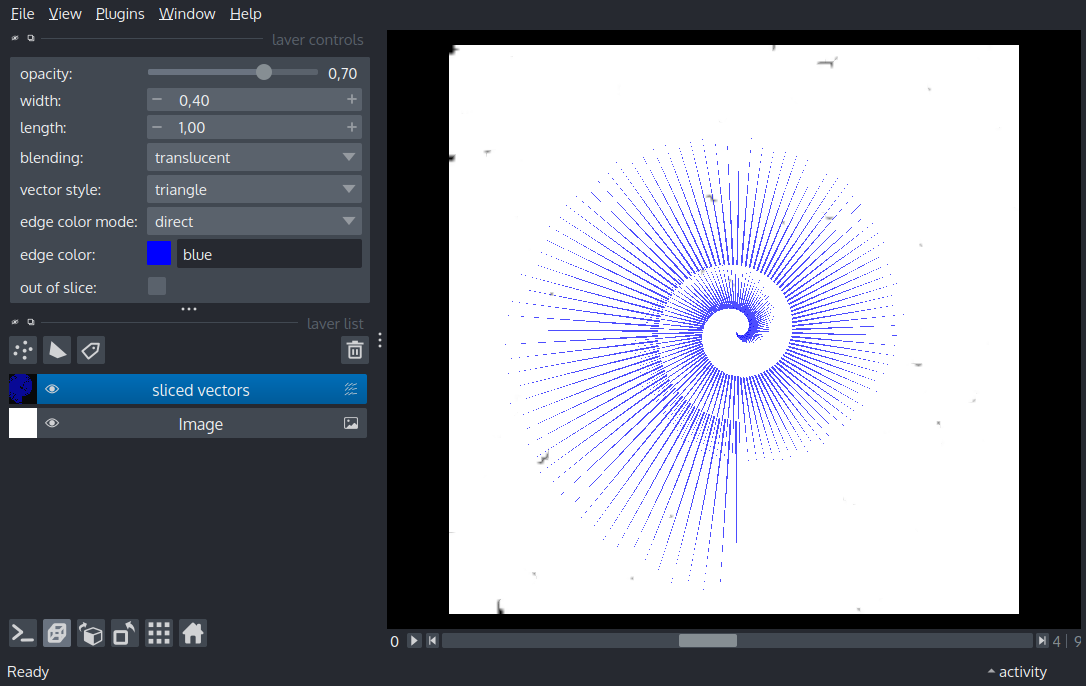
import numpy as np
from skimage import data
import napari
blobs = np.stack(
[
data.binary_blobs(
length=128, blob_size_fraction=0.05, n_dim=3, volume_fraction=f
)
for f in np.linspace(0.05, 0.5, 10)
],
axis=0,
)
viewer = napari.view_image(blobs.astype(float))
# sample vector coord-like data
n = 200
pos = np.zeros((n, 2, 2), dtype=np.float32)
phi_space = np.linspace(0, 4 * np.pi, n)
radius_space = np.linspace(0, 20, n)
# assign x-y position
pos[:, 0, 0] = radius_space * np.cos(phi_space) + 64
pos[:, 0, 1] = radius_space * np.sin(phi_space) + 64
# assign x-y projection
pos[:, 1, 0] = 2 * radius_space * np.cos(phi_space)
pos[:, 1, 1] = 2 * radius_space * np.sin(phi_space)
planes = np.round(np.linspace(0, 128, n)).astype(int)
planes = np.concatenate(
(planes.reshape((n, 1, 1)), np.zeros((n, 1, 1))), axis=1
)
vectors = np.concatenate((planes, pos), axis=2)
# add the sliced vectors
layer = viewer.add_vectors(
vectors, edge_width=0.4, name='sliced vectors', edge_color='blue'
)
viewer.dims.ndisplay = 3
if __name__ == '__main__':
napari.run()